🚀 We’re excited to share that our latest paper is out: Biologically Informed Procedure for Feature Summarization in Spatial Transcriptomics — a new framework designed to uncover the spatial organization of rare yet biologically crucial features, such as transcription factors, in imaging-based single-cell RNA sequencing data (e.g., MERFISH).
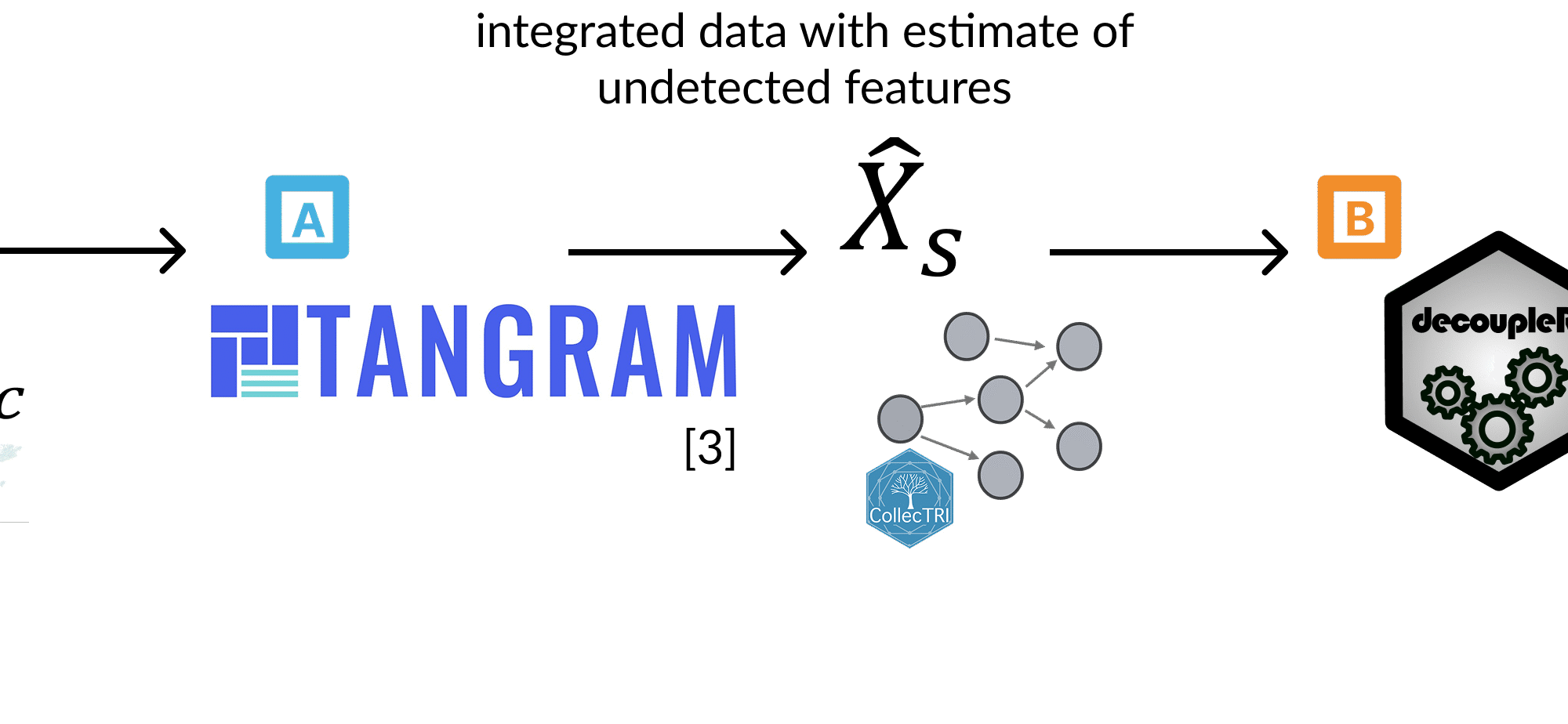
📊 In this work, we introduce a comprehensive procedure that integrates:
• TANGRAM, to align and integrate MERFISH with scRNA-seq data.
• Decoupler with COLLECTRI prior knowledge, to derive biologically grounded activity scores for transcription factors.
• Spatial statistics, to detect spatially informative features and highlight their organizational patterns across tissue sections.
🔍 By combining these elements, our workflow enables the discovery of subtle yet functionally relevant spatial signatures that would otherwise remain hidden — providing a powerful tool for interrogating tissue architecture in single-cell spatial genomics.
👥 This achievement was made possible thanks to the collaborative work of all authors and the support of the IJCNN organizers. Special thanks to the members of SysBioBiG – Systems Biology and Bioinformatics Group at UNIPD, who shared this journey with us.
🔗 The paper is available at: https://doi.org/10.1109/IJCNN64981.2025.11227921