On 16 January 2026, the “Guido Petter” Conference Hall at the School of Psychology hosted the annual meeting dedicated to the UNIPD’s activities within the DARE (DigitAl lifelong pRevEntion) initiative. The event featured an overview of the ongoing DARE projects led by UNIPD researchers, a seminar delivered by the Arsenal.IT consortium on the secondary use of healthcare data, and two roundtable discussions designed to encourage multidisciplinary exchange.
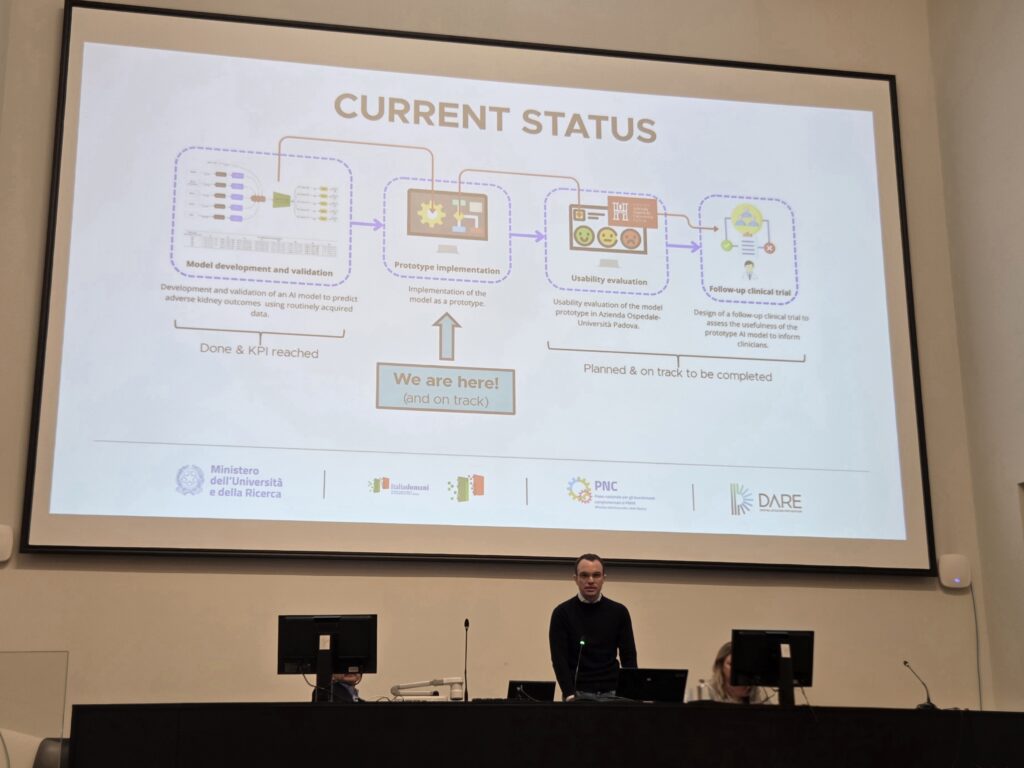
During the meeting, Professor Barbara Di Camillo took part in an insightful roundtable discussion entitled “From Research to Healthcare Practice: Working Together for New Perspectives in Prevention.” In addition, our researcher Enrico Longato presented the current progress of Work Package 3, Task 3.3a, entitled “Digitally-Empowered Management of Type 2 Diabetes: From Diagnosis to the Prediction of Complications.”
Overall, the meeting provided an important opportunity to exchange ideas, strengthen collaborations, and gain a comprehensive view of the diverse and innovative contributions of UNIPD within the DARE initiative.